The flu virus is a master of disguise. Researchers at IISc are racing to uncover its secrets.
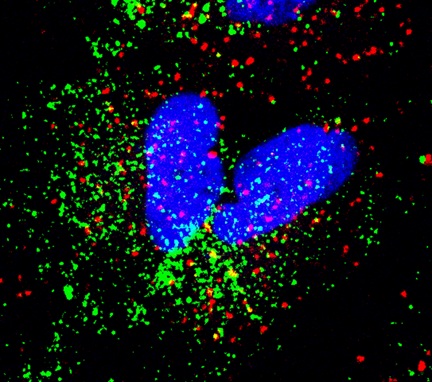
Red – Early Endosome Antigen 1 (EEA1), Green – IAV particles (Image courtesy: Shashank Tripathi)
In the winter of 1918, soldiers in cramped army barracks across Europe began falling ill with what looked like ordinary seasonal illness. Within weeks, many were dead – not from bullets, but from a virus that spread indiscriminately without warning. The misleadingly named “Spanish flu” pandemic went on to kill more people than World War I, resisting every medical countermeasure.
A grim sense of the virus’s reach even seeped into children’s rhymes:
I had a little bird
And its name was Enza
I opened the window
And in-flew-Enza
Influenza (flu) didn’t disappear after the war. In 1957, a new strain emerged in East Asia, then in Hong Kong in 1968, and in 2009, the H1N1 virus spread to every continent within weeks. Strains that have fully adapted to spreading in humans today (like H1N1 and H3N2) slipped into our species through these pandemics.
Like the faceless men from Game of Thrones, the flu virus can rapidly modify its appearance to escape our immune responses. These changes can be small mutations – called antigenic drift – or larger changes in viral proteins – called antigenic shift, which can produce an entirely new virus. The key proteins that drive these changes are two surface proteins called hemagglutinin (HA) and neuraminidase (NA), which can combine in different ways, giving rise to new strains.
Sometimes, these changes allow influenza to cross over to a new species altogether. A virus that once circulated only in birds could evolve to infect whales, seals, horses, or humans. Each new host offers the virus fresh chances to adapt. Earlier this year, that adaptability was on display in dairy farms in the USA, where veterinarians discovered the H5N1 bird flu virus in raw milk from cows. For a pathogen that usually sticks to birds, cows were a new territory and – worryingly – it appeared to be spreading from animal to animal.
But not every strain makes such a leap. Why is it that only some strains cross the species barrier, while others stay confined to their usual hosts? This is one of the mysteries that Kesavardana Sannula at the Department of Biochemistry is trying to unravel.
Tracking evolution
In recent years, Kesavardana’s lab has been focusing on a specific variant of the flu virus – the 2.3.4.4b clade of H5N1 (clade represents a group of viruses that have evolved from a common ancestor). This variant is rapidly acquiring mutations that allow it to thrive in a wide range of mammals. This is alarming because if H5N1 adapts to spreading among humans, scientists speculate that death rates could rise.

Kesavardana’s group has been trying to figure out what makes this clade different from previous H5N1 strains. What enables it to spill over to mammals from birds, and what kind of mutations is it picking up that might help it adapt to humans? In a recent study, his team found that mutations in certain viral proteins – especially in fox-adapted strains – could heighten the risk of human adaptability.
Another study from his lab uncovered a striking change in the neuraminidase (N) protein – a lengthier stalk – along with the loss of certain sugar groups on hemagglutinin (H). Together, these changes may influence how the virus binds to host cells and evades immune defences. His lab uses various computational structural biology methods, experimental validation, and collaborations with international influenza experts to track how these mutations affect viral fitness – the ability to cause disease and spread.
The H and N proteins can, in theory, pair in many different ways. Kesavardana’s group has shown that while many combinations can indeed occur, the virus’s fitness depends on precise molecular compatibility between the two proteins.
He is especially interested in why N1 (a subtype of neuraminidase) has been detected in past pandemic viruses and is exploring structural motifs that make it specifically concerning. These insights could have direct implication for global surveillance and vaccine design.
“The scary part is that this virus isn’t starting in one location like SARS-CoV-2 did,” says Kesavardana. “It’s already everywhere in wild birds and mammals. All it needs is the right combination of mutations to adapt to humans, and then it’s ready to spread globally.”
Staying ahead
Once the virus leaps to a new host, another question emerges: What happens inside the host, and how does the body respond? At the Centre for Infectious Disease Research, Shashank Tripathi’s group is exploring this tug-of-war between virus and host and finding ways to tip the balance in our favour.
The flu virus is a “forever virus,” Shashank says. “The range of host species it can infect is extensive. The potential for new flu strains to emerge is almost unlimited.”
Shashank’s group has found that H5N1 often triggers inflammatory cell death in the host that worsens tissue damage, while seasonal H1N1 tends to cause non-inflammatory death, leading to milder illness. Scientists can use such insights to develop host-directed antivirals – targeting pathways in host cells that the virus hijacks, so that even if the virus itself mutates, the treatment would still work. The idea is to combine these with drugs that act directly on the virus, slowing down its growth while also reducing harmful inflammation.

To find promising targets, Shashank’s team uses genome-wide CRISPR analysis, large-scale molecular data, and lab-grown tissue models to identify which host factors help the virus and which ones block it, in both H1 and H5 strains.
His lab is also working towards a flu vaccine designed to give long lasting and broad protection. It is based on a chimeric design combining HA – which triggers strong antibodies against both current and possible future variants – with a T cell component that targets conserved regions for lasting cellular immunity. This formulation can be delivered through a thermostable mRNA platform.
In parallel, his lab has tested an intranasal T cell vaccine in mice models that offered protection against H1, H3, and H5 strains – this could be paired with antibody-based vaccines for more effective protection. “Our ultimate goal is to make one vaccine for all [flu viruses], ideally a truly universal vaccine that offers good protection for at least 5-10 years,” adds Shashank.
Designing better vaccines
Flu vaccines have also been the focus of Raghavan Varadarajan’s lab at the Molecular Biophysics Unit (MBU). He and his team, in collaboration with the start-up Mynvax, which he co-founded, are preparing for clinical trials of a recombinant HA vaccine that has shown strong and broad immune responses in animal models and has also been found to be safe in early human trials.
Raghavan’s team uses structure-based design to present conserved sites in the HA stem more effectively to the immune system, eliciting a strong, targeted response. In parallel, his lab, in collaboration with Vidya Mangala Prasad’s group at MBU, is carrying out high-resolution structural studies to understand how certain antibodies neutralise the virus. They are also working on characterising the HA protein in complex with known neutralising antibodies in collaboration with Somnath Dutta’s group at MBU.

For Raghavan, however, the challenges aren’t just scientific. “People have to be aware and they have to be willing [to take flu shots and vaccines],” he says. Cost remains a barrier – flu shots are currently priced at around Rs 2,000, which is out of reach for many. Scaling up production and lowering prices, he adds, along with insurance or government coverage for vulnerable groups like pregnant women and children, could make vaccination far more accessible.
The biggest danger, however, remains the fact that even the most effective vaccines and antivirals may not be able to stop the virus, if conditions become conducive for its rapid spread. Shashank offers a sobering analogy: “When COVID-19 began, precautions [initially] worked. Then Omicron arrived, ten times more transmissible, and spread uncontrollably. If that happens with H5 – which has caused a recent outbreak in dairy cows in the USA – it will spread like a wildfire.”