Rahul Roy’s lab aims to engineer novel solutions for biological problems

Nine years ago, when Rahul Roy was applying to IISc for a faculty position, he gave presentations at several departments, because he wasn’t sure where he would fit in best. Although he was interested in both engineering and science, he didn’t foresee working in an engineering department. Eventually, however, he joined the Department of Chemical Engineering as an Assistant Professor, where he has been pursuing diverse projects at the intersection of engineering and biology.
Rahul’s lab, in his own words, is working towards the ambitious goal of making biology more “predictive”. Biological systems are inherently complex and heterogeneous. Take for instance the human body. Scientists still don’t know enough about how the different parts of our body work. This is because within each part, different cells behave differently. And within each cell, different molecules behave differently. This heterogeneity is a major problem in biology research; we cannot tell when a molecule will act differently.
This is also an issue when it comes to developing drugs and vaccines, which is now a painfully long process. Even after a molecule with therapeutic properties has been discovered, scientists need to test it first on cells in the lab, and then on animals, and finally perform clinical trials on humans to understand if they are safe and effective. But if researchers are able to better predict how our bodies would react to such treatments, it would be easier for them to bypass some of these steps and develop more effective and safer treatments in a shorter time.
Biologists with an engineering background like Rahul are using a combination of traditional biological techniques and new computational approaches to address such challenges. His lab is currently developing general “rules” that will help decrypt biological diversity at the molecular level and perhaps develop treatments without running clinical trials. “[While] we understand that there’s still going to be a large heterogeneity, we’ll still come up with rules which will explain the overall behaviour,” he says.
Towards this goal, Rahul’s lab is interested in using single-molecule imaging to study how different copies of the same molecule carry out the same function differently. Take the rate at which an enzyme catalyses a biological reaction, for instance. The value that scientists assign to it is merely the average of the rates for multiple copies of the same enzyme. Even little modifications to the enzyme’s structure can affect its catalytic activity drastically.
Another direction of his lab’s work is on the self-assembly of viruses and bacterial pore forming toxins. Understanding how these toxins assemble can help scientists find ways to block the bacteria from entering our cells and causing infections. A toxin found in some bacteria, called Cytolysin A, forms a 12-sided ring-like structure. In one of their collaborative studies, the group found that subunits of this toxin assemble into the ring in any random sequence and that the assembly is reversible — when incomplete structures are formed, some previously built structures can break up and release bits that join and complete the partial structures.
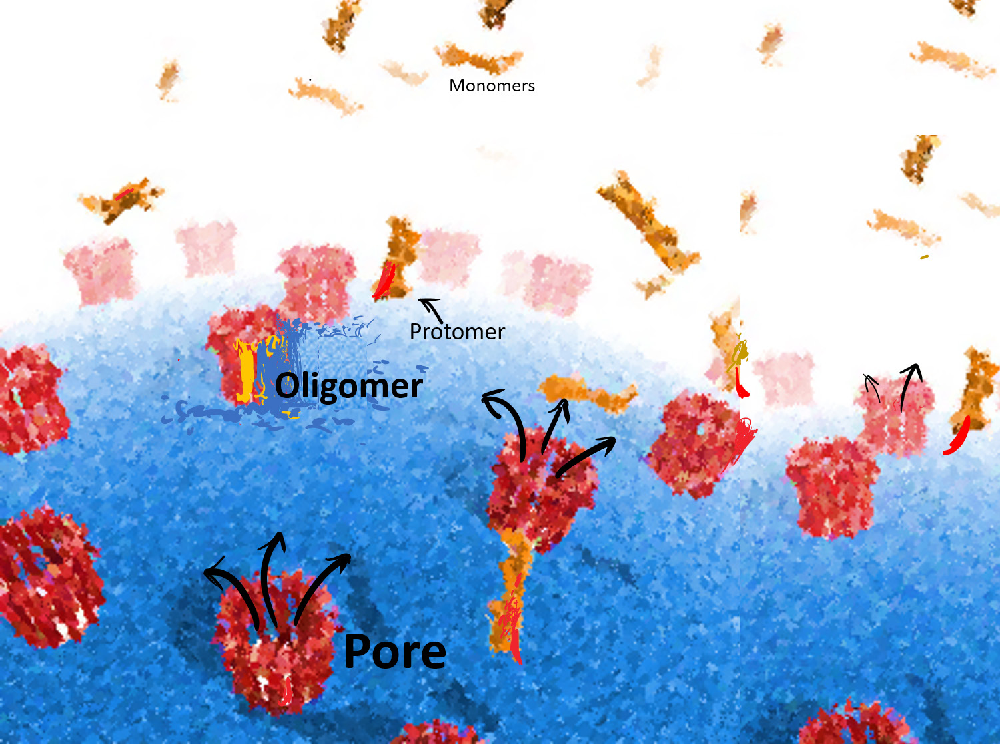
(Image courtesy: Satyaghosh Maurya)
A third area of interest is understanding how viruses like the novel coronavirus, SARS-CoV-2, mutate. In just the past few months, new SARS-CoV-2 variants have emerged. As the virus replicates, mutations arise and eventually accumulate in its genome. Errors during replication happen because RNA polymerase – the enzyme that creates copies of the virus – is inefficient. “Now, every time it [the virus] is making a copy of itself, it is actually creating a variant,” Rahul explains. This leads to the accumulation of multiple variants of the same virus inside a host cell, which requires the use of antivirals that can simultaneously act on all of them.
Using a technique called single virus sequencing, Rahul’s group has been able to discern the exact number of variants present in a blood sample. In another project, they have developed a method to count the number of RNA molecules that correspond to a particular gene in individual cells, by trapping a cell in a drop of a water-in-oil emulsion. This approach has helped them understand the exact concentration of a specific RNA molecule in each cell. Adding microfluidics to this approach allows them to study many cells simultaneously.
Yet another interest of Rahul’s lab involves building tools for sensing and lab-on-chip devices. His team has recently developed a smartphone-based antibiotic sensing approach, where sensors made up of DNA molecules, called DNA aptamers, are used to detect the presence of an antibiotic. When the sensor detects the antibiotic, dyes attached to it exhibit a change in either the fluorescence colour or intensity that can be easily detected by a smartphone camera. Such sensors can be designed to detect other kinds of chemicals as well. Rahul believes that, in the future, pluggable adapters for smartphones might help convert them into point-of-care diagnostic devices.
In recent months, Rahul’s lab has also been working on projects related to COVID-19. They have developed a rapid antibody test for COVID-19. A home-based test kit is also in the making, which aims to use antibody fragments to enrich and detect the virus concentration in easily available biofluids like saliva. It can further intensify the signal using an isothermal amplification technique at 37°C. This kit can display results within 20 to 30 minutes and can be used without professional assistance by anyone who is not able to visit a pathology lab.
(Photo courtesy: Satyaghosh Maurya)
Exploring multiple projects can be hard. Rahul explains that the reason why he ended up pursuing so many diverse projects is that he designs these with major inputs from his students and postdocs, which has often resulted in him pursuing areas that he never anticipated he would be working on, like microfluidics.
He says that his motivation to pursue research comes from the sheer joy of finding out something for the first time. Seeing the eyes of budding young scientists light up with joy when they understand a new concept also keeps him going. Students, being “naive”, often bring a unique perspective to a research problem because they do not carry the burden of preconceived notions, he says. “And that’s why we always want not one person, but hundreds of people approaching the same problem from different angles.”