In two recent studies, researchers at IISc and Unilever have collaborated to develop computational models of bacterial cell walls that can speed up the screening of antimicrobials – molecules which can kill disease-causing bacteria.
Each bacterial cell is enveloped by a cell membrane, which is in turn surrounded by a cell wall. Some bacteria like Escherichia coli (E. coli) are Gram-negative – their cell walls contain a layer of peptide-sugar complexes called peptidoglycans and an outer lipid membrane. Others such as Staphylococcus aureus (S. aureus) are Gram-positive – their cell walls only have several layers of peptidoglycans.
Antimicrobials kill bacteria either by disrupting the cell wall’s lipid membrane and destabilising the peptidoglycan layer, or by translocating through the cell wall layers and disrupting the cell membrane inside. However, the actual mechanisms of interaction between antimicrobial molecules and these cellular barriers are poorly understood. “The cell envelope is a big part of this puzzle, and it is often overlooked,” says Pradyumn Sharma, a former PhD student at the Department of Chemical Engineering (CE), IISc, and one of the authors.
In one study, the team created an ‘atomistic model’, a computer simulation that recreates the structure of the cell wall down to the level of individual atoms. They incorporated parameters such as the sizes of sugar chains in the peptidoglycans, the orientation of peptides, and the distribution of void size.
“The structure of the peptidoglycan layer is semi-permeable, because nutrients and proteins that bacteria need, have to pass through,” explains Ganapathy Ayappa, Professor at CE and corresponding author. These are the same voids that the antimicrobials also pass through. Rakesh Vaiwala, a Research Associate at CE and one of the authors, adds that their team is the first to propose a comprehensive molecular model of the cell wall for S. aureus.
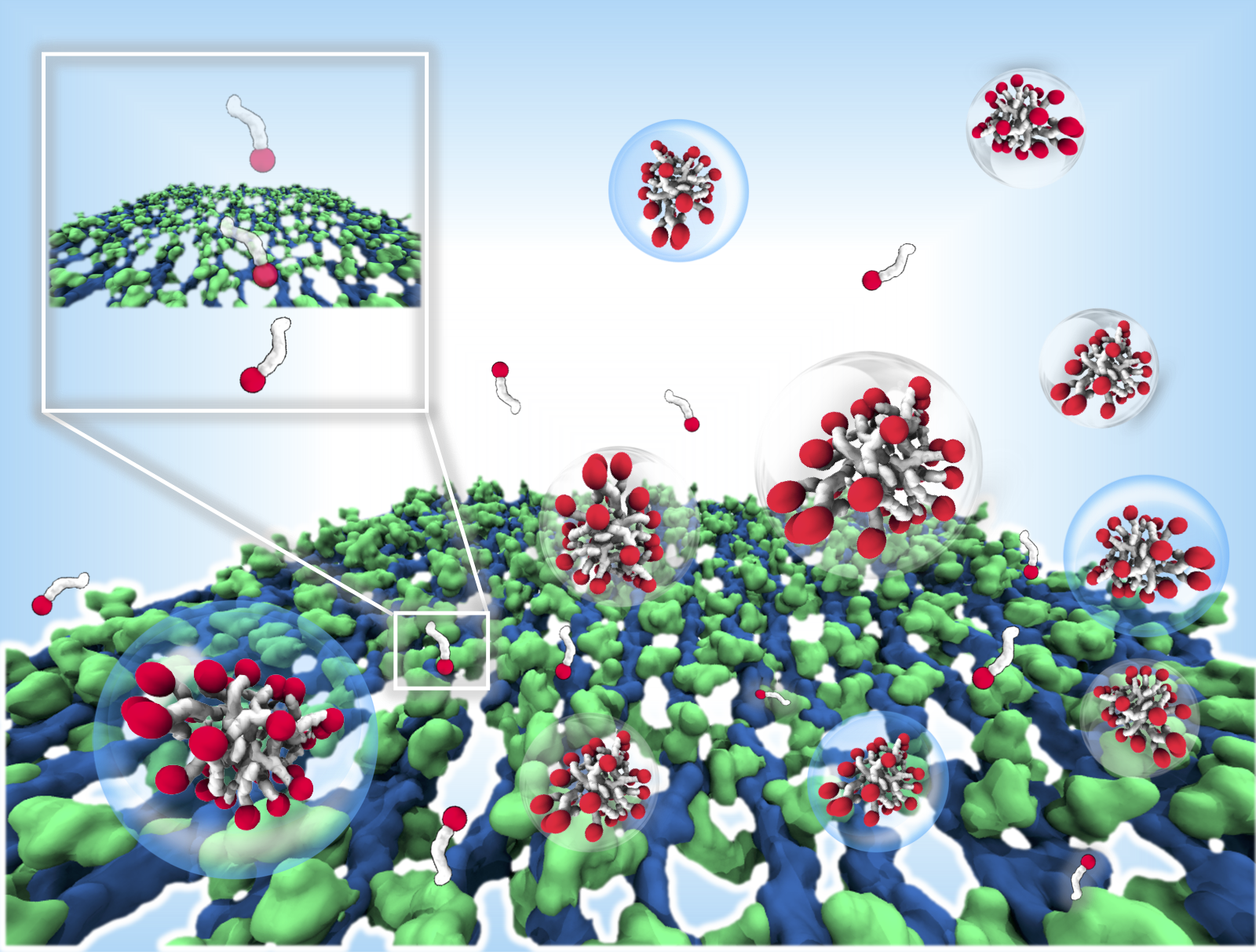
Using the supercomputing facility at IISc, the team tested the effectiveness of their model with several known antimicrobials. One of these, melittin, a short peptide, binds with higher efficiency to the E. coli cell wall than that of S. aureus. The researchers found that melittin interacts with peptides involved in a process called transpeptidation in peptidoglycan biosynthesis, and can potentially disrupt cell wall integrity. Thymol, a naturally occurring small molecule, translocated rapidly through the whole stack of peptidoglycans in the cell wall of S. aureus.
In the other study, the team used their model to compare the movement of different surfactant molecules through the peptidoglycan layer in E. coli. Like detergents, surfactants have a water-loving ‘head’ attached to a water-avoiding ‘tail’ chain. The team showed for the first time the link between the length of the tail and antimicrobial efficacy of surfactants. Surfactants like laurate with shorter chains translocated more efficiently than longer chain oleate. This was corroborated by experiments carried out by scientists in the Unilever team, which showed that shorter chain surfactants killed bacteria at a higher rate than surfactants with longer chains.
The team also collaborated with Jaydeep Kumar Basu, Professor in the Department of Physics, to create vesicles composed of E. coli extract and observed their interaction with surfactants under a microscope. The vesicles were found to burst open at a much faster in the presence of laurate compared to oleate.
“The goal with Unilever is to facilitate rapid screening of molecules using the computational models we have developed, to narrow down the search for potential antimicrobials to a smaller subset of molecules which can be tested in the laboratory,” explains Ganapathy Ayappa.