Researchers at IISc have designed a short peptide capable of poisoning a key enzyme in disease-causing bacteria, including some of most deadly and antibiotic-resistant species.
Made from a short stretch of about 24 amino acids, the peptide mimics the action of a natural toxin which inhibits a class of enzymes called topoisomerases. These enzymes play a crucial role in unspooling and re-coiling bacterial DNA during replication and protein synthesis. They are an attractive target for antibiotics because the ones in bacteria are very different from those in humans.
Among the most widely used antibiotics are fluoroquinolones such as ciprofloxacin, which target these topoisomerases. However, overuse of these antibiotics around the world has led to the alarming rise of antibiotic-resistant bacteria, prompting scientists to pursue alternative strategies and molecules.
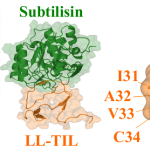
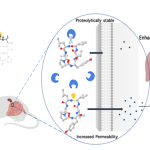
Topoisomerases form a covalent adduct – an intermediate complex – with the bacterial DNA, to coil or uncoil it. The peptide developed by the IISc team binds to this adduct and “traps” it in place, kicking off a cascade of events that lead to cell death, explains Raghavan Varadarajan, Professor at the Molecular Biophysics Unit (MBU), and one of the corresponding authors of the study published in EMBO Reports. This is similar to how a natural toxin called CcdB, produced by certain other bacteria and plasmids, works.
“The full length CcdB protein is large. It is not feasible to use it as a drug in its entirety,” says first author Jayantika Bhowmick, former PhD student at MBU and currently a postdoctoral researcher at the University of Cambridge. Instead, the team snipped out a small stretch from the tail end of this protein and added a few more amino acids that would allow the new peptide to enter bacterial cells. The peptide design was carried out by the lab of Jayanta Chatterjee, Professor in MBU.
The team then tested the new peptide’s effect on the growth of several disease-causing bacterial species, including E. coli, Salmonella Typhimurium, Staphylococcus aureus and a multidrug resistant strain of Acinetobacter baumanii – both in cell culture as well as animal models, in collaboration with the lab of Dipshikha Chakravortty, Professor at the Department of Microbiology and Cell Biology (MCB). They also compared the effect of their peptide against clinical doses of ciprofloxacin. Depending on the species, the peptide was found to either block or “poison” a specific type of topoisomerase – an enzyme called DNA gyrase in many of them, explains Manish Nag, PhD student at MBU and another author. “It is [also] capable of disrupting most of the strains’ membranes,” he adds.
In animal models, the peptide was found to drastically reduce infection. “In most of the cases, we saw that the decline in the bacterial count in major organs following peptide treatment was higher than in the ciprofloxacin-treated group. That was pretty encouraging to us,” says Bhowmick. For example, in animals infected with antibiotic-resistant Acinetabacter baumannii, the peptide treatment caused an 18-fold reduction in bacterial load in the liver, compared to only a 3-fold reduction by ciprofloxacin. The peptide was also found to be relatively safe and did not cause toxic reactions in the animals.
Since the peptide binds to a different site on the bacterial enzyme than ciprofloxacin, the researchers believe that it provides leads for identification of drugs that can be used as a combination therapy with existing antibiotics. Varadarajan adds that the study also reinforces the importance of targeting topoisomerases as a valid approach to finding new antibiotics.