Faced with the growing threat of antibiotic resistance, researchers at IISc and elsewhere are in pursuit of alternative strategies
In the early 1940s, as World War II ravaged several parts of the globe, great uncertainty and a sense of doom loomed large over the soldiers and their families. Even a tiny splinter on the battlefield could prove fatal due to the resulting infection and gangrene, for which effective treatment was scarce. But then, a beacon of hope emerged – a ‘wonder drug’ called penicillin, Alexander Fleming’s accidental discovery. The subsequent mass production of penicillin was instrumental in ushering in the golden era of antibiotics, a class of drugs that has collectively saved millions of lives since then. However, nearly a century later, rising antibiotic resistance threatens to take these efforts back to square one.
Antibiotic resistance is when bacteria develop the ability to block the action of antibiotics – molecules designed to kill them or slow down their growth. This makes bacterial infections harder to treat, and routine procedures like surgeries and transplants risky. The World Health Organisation (WHO) estimates that every year, an alarming 2,30,000 people die worldwide as a result of multidrug-resistant tuberculosis alone – this figure is much higher if we consider deaths due to other resistant species of bacteria. Even simple skin infections are becoming increasingly difficult to treat, owing to the emergence of strains that are resistant to most known antibiotics.
Given the gravity of the situation, scientists around the world are racing against time to develop more effective antibiotics as well as alternative approaches. Bacteria are adept at developing resistance to drugs that block specific pathways or processes by making small changes to the targeted pathways or molecules. To counter this, scientists have been developing molecules that directly bore holes in the bacterial cell membrane. Rupturing the membrane would disable all essential processes almost immediately, making it difficult for the bacterium to develop resistance. Researchers both at IISc and elsewhere have been developing novel antimicrobial peptides (AMPs) and nanoparticle-based artificial enzymes to accomplish just this.
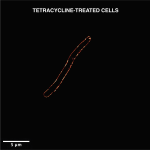
Another exciting development is the possibility of exploiting the existing prey-predator relationship between bacteria and bacteriophages – viruses that can kill bacteria – to develop new antimicrobials. “Phages were used quite a bit in the early 1900s on the research scale and also on humans. But with the discovery of penicillin and other antibiotics, [they] fell off the way,” says Rachit Agarwal, Assistant Professor at the Centre for BioSystems Science and Engineering, IISc. These bacteriophages – or phages – enter bacterial cells, multiply, and then burst the bacterial cell open to release the new phages, thereby killing the bacterium.
Rachit points out that while there are only a few classes of antibiotics, the repertoire of phages is much larger, and these phages target many different receptors on the bacterial cell membrane – giving us a wider range of options to choose from for therapy. His lab recently tested the use of a mixture of five different phages to treat mycobacterial infections. Mycobacteria are notorious for their ability to survive in extreme conditions of low oxygen or pH inside our body, and they can stay dormant without dividing for long periods of time, according to Rachit. “We were actually quite surprised to see, at least from whatever lab testing we did, that these phages were quite effective against mycobacteria in all of these pathophysiological environments that we looked at.” When his team combined phage therapy with antibiotics, they found that the ability of the phages to kill the bacteria improved significantly.
Since phages are large molecules, delivering them directly into the niches that bacteria occupy inside our body could make therapy more effective, according to Rachit. Mycobacteria hide within our cells and therefore delivering phages inside these is an additional challenge – this is something that his lab is actively pursuing. “Right now, we’re mostly focused on using some polymeric carriers to load the phages, and modifying these carriers either in size, charge or surface properties to be able to load phages, get internalised by mammalian cells, and be transported to the right intracellular locations,” he says.
(Image: Arunabh Athreya)
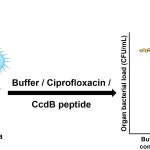
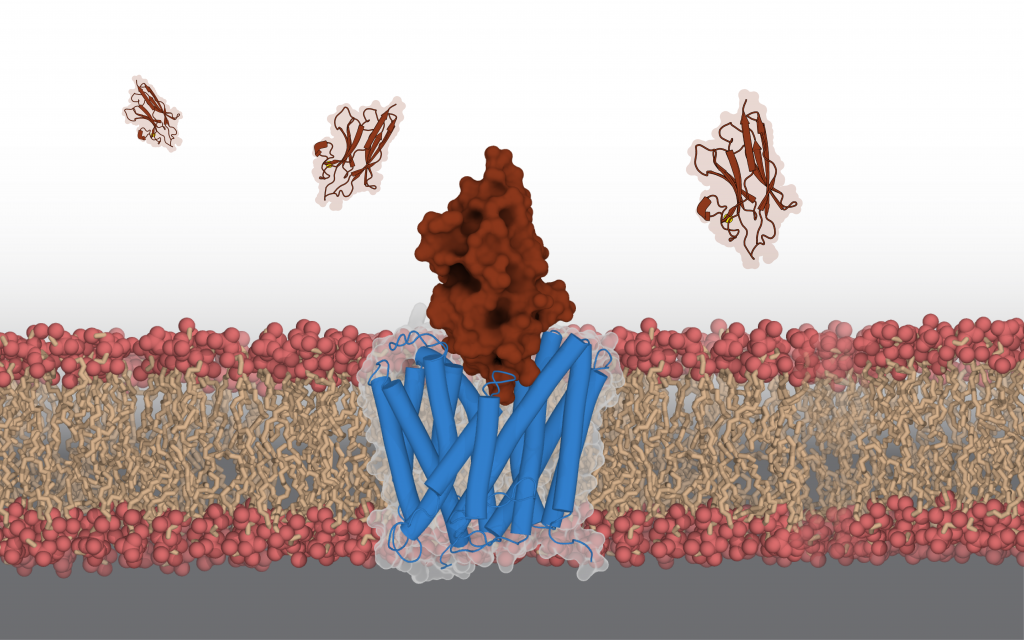
Aravind Penmatsa, Assistant Professor at the Molecular Biophysics Unit, IISc, has a different approach to this problem of antibiotic resistance. His lab explores the possibility of blocking certain types of transporter proteins on the bacterial membrane that help it become resistant. These proteins help transport nutrients and other essential molecules. “Some of these transporters are used by the bacteria … to pump out antibiotics or antibacterial compounds that we use on a day-to-day basis into the external environment. So, this becomes a major mechanism for gaining antimicrobial resistance,” says Aravind. “If you can somehow control this efflux process, and block it, the potential for this strategy is that you can enhance the efficacy of existing antibiotics.”
Aravind’s lab has exploited this principle by developing antibodies derived from mammals like camels which can block a specific efflux pump in Staphylococcus aureus, a notoriously resistant species of bacteria. These antibodies are different from conventional Y-shaped antibodies found in humans (and several other species) in that they have a single domain for recognition of the target protein, and are shaped like “a rugby ball,” Aravind explains. This property allows them to interact with crevices of target proteins more effectively, making them an interesting candidate for developing efflux-pump blockers. However, harnessing this technology to treat bacterial infections will take time, and will require one to choose the right targets, he says. “These single-domain camelid antibodies are phenomenal in their ability to specifically detect the presence of efflux pumps. So even if the whole idea of a therapeutic blocker will take a long time, the idea of using them as diagnostic tools is a much more real and practical application for these antibodies,” he adds.
Although several such efforts have gone into tackling existing antibiotic-resistant strains, preventing the rise of newer antibiotic-resistant species also rests heavily on human behaviour. “A lot of this antibiotic resistance is also due to patients not completing their [therapy] regimen,” says Rachit. “So, if somehow patient compliance can be increased, then the expectation is that the evolution of antibiotic resistance would slow down.”